The Collaborative Cross Graphical Genome (CCGG) is a graph-based pangenome representation for the widely-used recombinant-inbred mouse genetic reference population known as the Collaborative Cross (CC). This version was constructed by merging the standard mouse reference, GRCm38, with de novo assemblies of the 7 founder's genomes guided by the short-read sequence data from CC-line samples. It packs 83 genomes in a single graph which directly captures important notions relating genomes such as identity-by-descent between mouse strains, and highly variable genomic regions. The introduction of special anchor nodes with sequence data provides a coordinate framework that divides all genomes into homologous segments. Parallel edges between anchors contain all genomic variants and indicate shared haplotypes. Furthermore, genomic annotations are provided that include gene and exon intervals, repeat types, and the of alignments edges relative to the mouse reference assembly. The graphical structure of the CCGG with annotations provides a comprehensive picture of the genome structure surrounding biological features, which are easy to traverse for searching, annotating, comparing and visualizing features. Integrating sequencing data with CCGG, the recombination hotspots, especially within the gene regions, are identified in CC strains. THe CCGG provides an effective pangenome model that represents the genetic diversity of the CC and facilitates tool chain development for downstream analysis.

CCGG
Collaborative Cross Graphical Genome
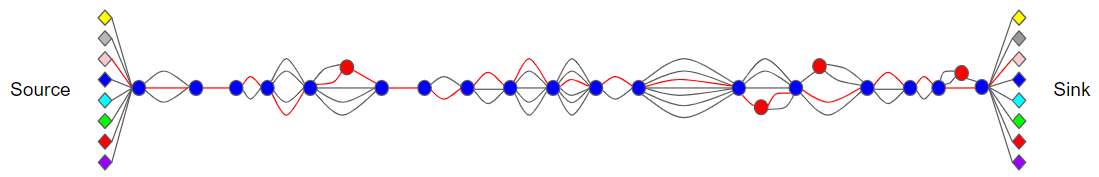
The CCGG pangenome is a single directed, k-partite graph composed of nodes and edges.
Node types
Anchor nodes, represented as blue circles, are conserved, unique 45mers with consistent genomic ordering in all genome assemblies. The sequence of each anchor node is supported by multiple reads in every sequenced CC sample.
Source and Sink nodes, represented as diamonds above, contain no sequence or annotations and represent the start and end of each assembled contig.
Floating nodes, represented as red circles, contain no sequence or annotation and only appear as source (src) or destinations (dst) in edge annotations. Inserting floating nodes between shared subsequences allows for sequence sharing (compression) between anchor pairs.
Edges
Edges represent sequences between nodes in the graph and contain all the sequence diversity. Each edge is annotated according to the one or more genome assemblies upon which it lays. We consider two or more edges that share common source and destination nodes as parallel. This notion of parallel extends to subgraphs that are parallel to paths separated by any pair of anchor nodes.
Path
A series of nodes and edges sharing common source and destination anchors. A genome contig can be represented as a Path between a “SOURCE” and “SINK” node.
Reference
The Collaborative Cross Graphical Genome
Hang Su, Ziwei Chen, Jaytheert Rao, Maya Najarian, John Shorter, Fernando Pardo Manuel de Villena, Leonard McMillan
bioRxiv 858142; doi: https://doi.org/10.1101/858142
If you have any question or comment, please contact at ziwei75@live.unc.edu or hangsu@email.unc.edu.